Viruses are infamous for using our own system against us. Most viruses do that, however individual viruses have their own strategies in how they will achieve it. The story for any virus begins with attaching to a receptor on our cells and then gaining entry. Once they enter they multiply and make proteins that are structurally and functionally important for the virus. They assemble into new virions and exit the cells to infect nearby cells or shed from our body to a new host.
Illustrated by @scienceblocks using following images -
Crime by Aby Badali | CC BY-SA 2.5
SARS-CoV-2 by CDC/ Alissa Eckert, MS; Dan Higgins, MAM | Public Domain. Enterocyte by Boumphreyfr | CC BY-SA 3.0
The life cycle of SARS-CoV-2
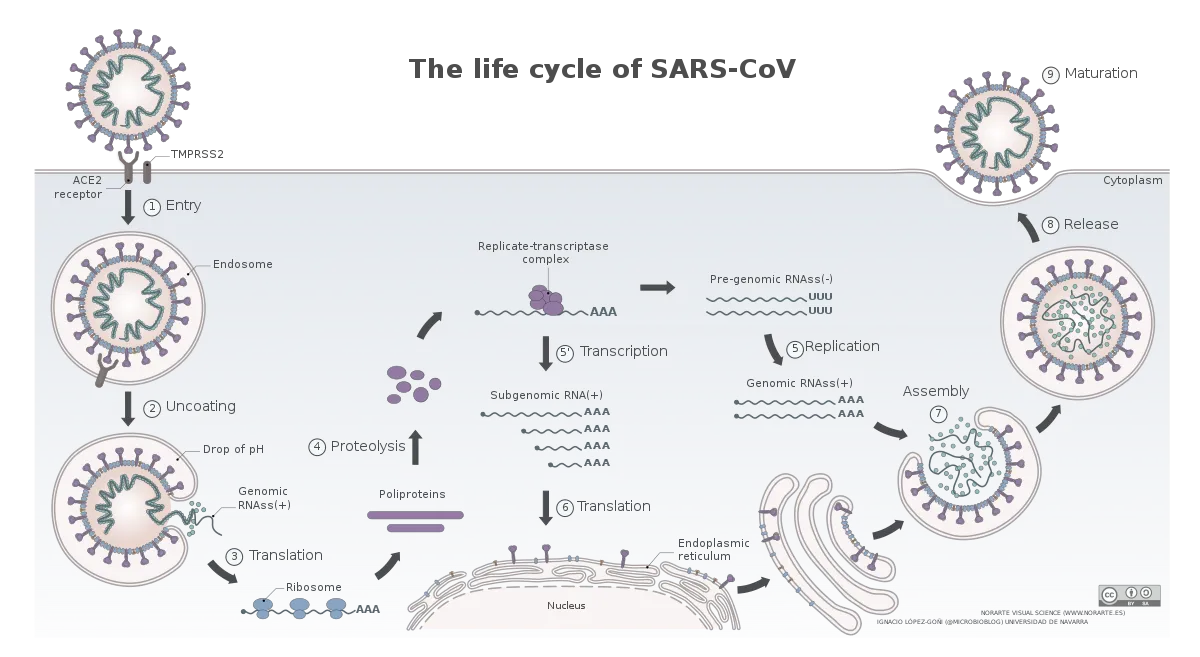
Image by Vega Asensio | CC BY-SA 4.0
The novel coronavirus, SARS-CoV-2, have a similar life cycle. Once, it enters the respiratory tract it binds to angiotensin-converting enzyme 2 (ACE2) receptor on the surface of some cells (Hoffman et al., 2020). This is followed by cleavage of spike protein on the surface of the virus via a protease TMPRSS2. This exposes a domain of spike which promotes membrane fusion between the virus and the cell. The virus hence gains entry into the cells and gets on with its business. However, which cells in the respiratory tract and other organs known to be infected by this virus express ACE2 and how is the expression of ACE2 regulated in these cells is not fully understood.
Check out this video for detailed explanation.
The paper by Zeigler et al., 2020, that we will discuss today gives us a hint about exactly that. It talks about which cells where express ACE2 and TMPRSS2, and more importantly how the expression of ACE2 might be regulated. It also gives us a hint that how the SARS-CoV-2 might actually be using the antiviral response of the cells to its own benefit. And if that is so what can this regulation tell us about who is more likely to be severely affected? What about using antiviral interferon as therapy? Which model of the disease we should or should not use when studying the disease or testing new cures?
Which cells express ACE2?
Zeigler et al., took a very interesting approach to figure out which cells express ACE2 and TMPRSS2. They have analysed the single cell RNA seq (scRNAseq) data from lungs, nasal passage and gut of humans and non-human primates. The cool thing about scRNAseq is that it looks at the genes expressed in individual cells and create clusters of cells, such that cells with similar overall gene expression fall into one cluster. Then using the known genes which are expressed only particular kind of cells you can identify what cell type a particular cluster correspond to. You may then ask the question that in which of these clusters your gene of interest is expressed in.
The cells in nasal passage and lungs that CoV-2 would love to infect
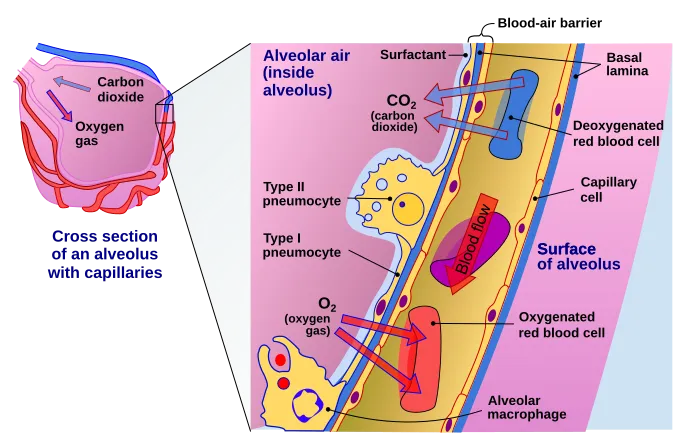
Image by Delmalani18 | [CC BY-SA 4.0]
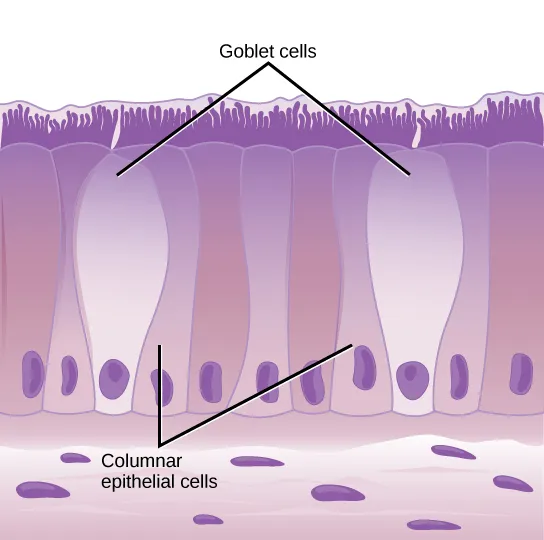
Image by CNX OpenStax | CC BY 4.0
In the lungs, the ACE2 expression was majorly found in type II pneumocytes and in ciliated cells of respiratory tract epithelium. To give you an idea of numbers about 1.4% type II pneumocytes, and 7% ciliated cells expressed ACE2. 0.8% of Type II pneumocytes and 5.3% of ciliated cells expressed both ACE2 and TMPRSS2. But that's the story of the lungs. However, if the virus has to infect it first needs to attach and infect cells close to the entry point. They identified that secretory goblet cells were the major target for SARS-CoV-2 in nasal epithelium.
The gut has target cells for CoV-2, as well
Now, if you are aware of symptoms of Covid19, the disease caused by SARS-CoV-2, you might know that it is characterised by pneumonia, fever, cough and sometimes even diarrhoea. Moreover, it has been shown that covid19 patients keep shedding the virus in stool for quite some time (Wu et al., 2020). The virus has also been shown to infect the GI tract (Xiao et al., 2020, Wang et al., 2020). Xaio et al., found that ACE2 was majorly expressed in cilia of glandular epithelium and they found the viral protein in stomach, duodenum and rectum. In line with this Zeigler et al., identified ACE2 expressing cells in the ileum of the small intestine - absorptive enterocytes.
The regulation of ACE2 by anti-viral molecules - Interferons.
ACE2 - interferon-stimulated gene?
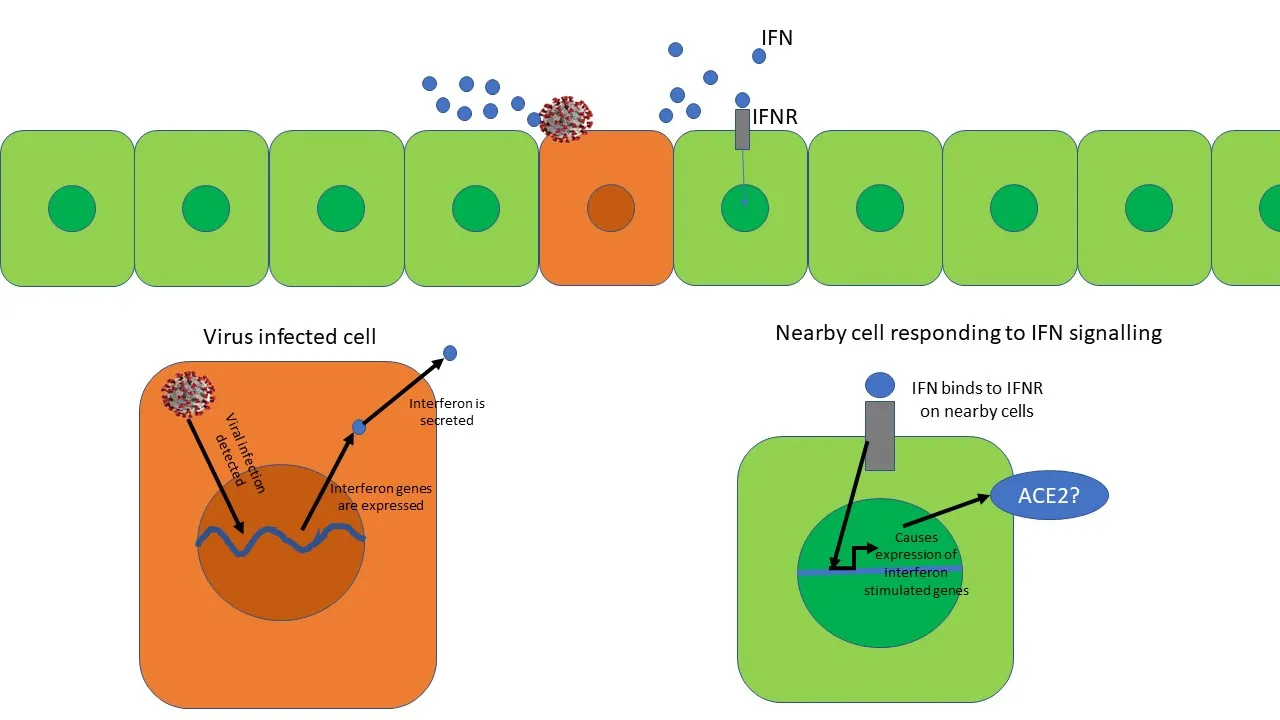
Anti-viral interferon response. The virus infected cell secretes interferon's (IFNs). The IFN binds to interferon receptor (IFNR) on nearby cells and starts expression of interferon-stimulated genes. However, is Ace2 an interferon-stimulated gene? Illustrated by @scienceblocks
There was something more interesting going on here. No matter where the authors took samples from - non-human primates (Rhesus macaque) or humans, they found that expression of ACE2 always correlated with expression of genes known to be expressed during an interferon response - a cellular defence against viral infection. Now, this is interesting, because interferons are class of molecules released by virus-infected cells to warn the nearby cells to up-regulate their viral defences. They also modulate the immune response and increase expression of MHC on cells (a molecule that displays viral peptides to T cells).
To confirm that interferon can indeed increase the expression of ACE2 in cells, Zeigler et al., treated cells derived from human nasal epithelium with interferon-gamma and interferon-alpha 2. They noticed a drastic increase in the expression of ACE2 gene in these cells. So, what does it mean if an anti-viral response of interferon can itself increase the receptor to which the virus will bind on the cells? Does it mean that the SARS-CoV-2 would use our viral defence system against itself? Is the virus being like - bring that interferon on humans! it may slow me down a bit, maybe, but it will also increase my chances to infect nearby cells? Moreover, given this information what does it mean for the interferon therapy proposed as a possible treatment against the virus? Is it really a good idea?
Viral infections increase the expression of ACE2?
That being said, what does interferon increasing expression of ACE2 mean for people with other viral infections. It is likely that people infected with any other virus may have a higher expression of ACE2 in certain cells and could be at higher risk of contracting Covid19 and becoming more severely ill? At least, in this paper, the authors show that HIV+ patients have higher expression of ACE2 in their cells. Given that HIV infection is known to cause a chronic interferon response in the body, it may explain why. Moreover, infecting non-human primates with Simian-Human immunodeficiency virus (SHIV) causes an increase in ACE2 expression in their cells as well. But, HIV and SHIV are not the only viruses that cause an interferon response and hence an increase in ACE2 expression. Humans infected with influenza A or B virus shows an increase in ACE2 in their cells. And it was not just the viral infected cells that showed an increase in expression of ACE2 but non-infected nearby cells showed this increase too.
ACE2 expression regulation in other diseases?
This brings me to yet another question. What about people with other non-viral co-morbidity? Can the interferon related increase of ACE2 explain some of the severity of the disease in these people? It is a good question to address. Well, at least a transcriptomic analysis of the lungs of patients with co-morbidities associated with covid19 show higher expression of ACE2 in their lungs (Bruna et al., 2020). But, it remains to be seen in which conditions this increase in ACE2 expression dependent on interferon at all. Maybe, we can have more discussion on it in the comment section.
Cell lines and mouse models
All that said and done, here is one more take-home message that comes out of this study by Zeigler et al., 2020. This is regarding the appropriate cell lines and animal model. For instance, in this paper, they were not able to find any increase in ACE2 expression in a bronchial epithelium - BEAS 2B cell line, on stimulation with interferon. However, they did see this happen in patient-derived cells. Well, cell lines are often transformed and change of the course of cell culture conditions. It happens many times that cell lines don't represent what happens in actual cells. And, patient-derived cells primary cells in culture are much more closer to their in vivo counterparts than cell lines. In fact, BEAS 2B cell line does not express ACE2, to begin with.
In fact, in this regard, I would like to mention this paper by Hoffmann et al., 2020 which can serve as a good guide for which cell line to use. A good human cell line as far as infection by SARS-CoV-2 is concerned would be Caco 2 (human epithelial colorectal adenocarcinoma cell line) or Calu 3. (lung cancer cell line). Vero E6, a cell line derived from African green monkey kidney epithelial cells, also serves as a good system. Moreover, it would be interesting to see if SARS-CoV-2 itself can induce an interferon response that increases ACE2 expression in these cell lines or in primary cells. And if that happens the inhibition of infection in, in vitro studies using cell lines that don't respond to interferon in a similar way may not produce an accurate picture. However, these cell lines would nevertheless be fine in screening for drugs that inhibit Spike-ACE2 binding, for instance.
Moreover, the authors found that mouse tracheal epithelial cells do not respond to interferon as human cells do. Interferon increased the expression of other interferon-stimulated genes in mouse cells but ACE2 should only a slight increase, Same was true for mouse treated with intranasal interferon spray. There was only a slight increase in the number of cells expression ACE2 and a slight elevation in the expression of ACE2 gene. So, this again makes one ask whether a mouse model infected with SARS-CoV-2 paint the exact picture for certain studies. For example, would an animal trial with interferon-gamma or interferon-alpha 2 in combating Covid19 a good model?
Anyhow, the regular mice itself is not a great model for SARS-CoV-2 by itself. There is a difference in the sequence of human and mouse ACE2 protein and hence Spike of SARS-CoV-2 doesn't bind as efficiently with mouse ACE2 as it does with Human ACE2 (see the link). Nevertheless, there is a transgenic mouse which comes to the rescue - K18hACE2 mice. This mouse is genetically modified to express the human version of ACE2 in airway epithelial cells (McCray et al., 2006). Since this mouse already has a high expression of human ACE2 in its airway, it is a good model to understand drugs that can inhibit viral binding and entry into the cells. It will even work fine for other downstream processes like viral replication inhibitors and viral protease inhibitors. However, when it comes to biologicals such as interferons or those that modulate the immune system to indirectly affect the virus, it must be better to evaluate that how much this mouse model mimics what happens in humans in response to a similar drug.
Final remarks
I think what authors found in this paper is definitely worth digging into further. One drawback of paper is that it mostly looks at RNA level of ACE2, but doesn't dwell much into questioning about protein levels of ACE2 in the cells of interest (Except one in vitro experiment). Moreover, I would love to know how interferon regulates ACE2. They mention that STAT1 might be regulating the expression of ACE2 in response to interferon. However, if that is the case then why this does not happen in mice? Is the promoter sequence for ACE2 different in humans vs mice? That is something someone needs to work on. Finally, I would like to know how ACE2 is regulated in Covid19 related comorbidities - Type 2 diabetes, hypertension, cardiovascular diseases, cancer, etc?
Anyhow, I guess that is it for today. We can continue the discussion in comments. Do share whatever comes to your mind.
About STEMSocial
But, before I go I would like to mention about the STEMSocial platform. Well, if you love reading and writing interesting science articles @steemstem or now STEMSocial is a community on hive that support authors and content creators in the STEM field. If you wish to support @steemstem do see the links below.
Or delegate to @steemstem.
25HP
50HP
100HP
200HP
250HP
500HP
1000HP
10000HP
100000HP
Also, if you have any questions regarding steemstem, do join the steemstem discord server.
You can DM me on discord, I have the same handle - @scienceblocks. Also if you are not a steem user, and reading this blog inspired you to start your science blog, find me on discord and let me know about you. I can try and help you navigate your way through steem.

Video by @gtg
References
Hwang et al., 2018. Single-cell RNA sequencing technologies and bioinformatics pipelines
Wu et al., 2020. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples
Xiao et al., 2020. Evidence for Gastrointestinal Infection of SARS-CoV-2.
Wang et al., 2020. Detection of SARS-CoV-2 in Different Types of Clinical Specimens
Borden, Silverman and Sen. Chapter 52 - Interferons
Mice, hamsters, ferrets, monkeys. Which lab animals can help defeat the new coronavirus?