
@sco is doing a "my shittiest method" challenge thing and I have been nominated by @scienceangel ! Took me some time but now I am writing it. Although it's more of a "my shittiest experiment" than "my shittiest method", as the method was technically fine.
The rules for this
- describe the worst method/assay/experiment you have to work
- use the #myshittiestmethod and the #steemstem tag
- use “My shittiest assay method: … ” as title
- you can nominate some peers, they have to participate then (unless they want to be seen as very, very lame). But you can join the party without invitation as well!
So, here we go.
Some Background
At the time, I had a 7 weeks internship in a microbiology lab (about which I wrote one of my first posts, the only post of mine ever to be curied!), where they were researching specific protein channels in the membrane of the ER.
What?
The ER (or Endoplasmic Reticulum) is a large part of eukaryotic cells - cells that have a nucleus (so no bacteria!). It has several functions, making sure proteins are folded correctly is one of them because proteins usually need a 3-dimensional structure to function as intended.
On the membrane of the ER, there are several channels that are responsible for transporting proteins into and out of the ER, because simple diffusion would be way too slow. In yeast, one of these channels is called SEC61.
The question in my experiment was, if the overexpression (= "cell uses a specific gene more than usual to produce a specific protein more than usual") of specific proteins has an effect on the export out of the ER when certain protein channels don't work anymore.@suesa
Why?
Well, nobody is 100% sure how much SEC61 and other channels are involved in said export.
But why do we care?
In humans, a similar channel in the ER-membrane exists. Proteins that are wrongly folded in the ER (it's a biological system, there are always mistakes) need to be transported to the outside and taken apart again, or the cell might die.
There are a lot of diseases, especially neurodegenerative diseases like Alzheimer's disease are linked to the aggregation of misfolded proteins. Knowing how the cell transports them and how to support or fix this system is thus something very interesting.
And it's easier to test the basics in yeast, before doing weird stuff to humans, you know?
The Experiment
The insertion of the genes to overexpress the proteins we wanted to check into the "damaged" yeast strains was a piece of cake and worked without any further issues.
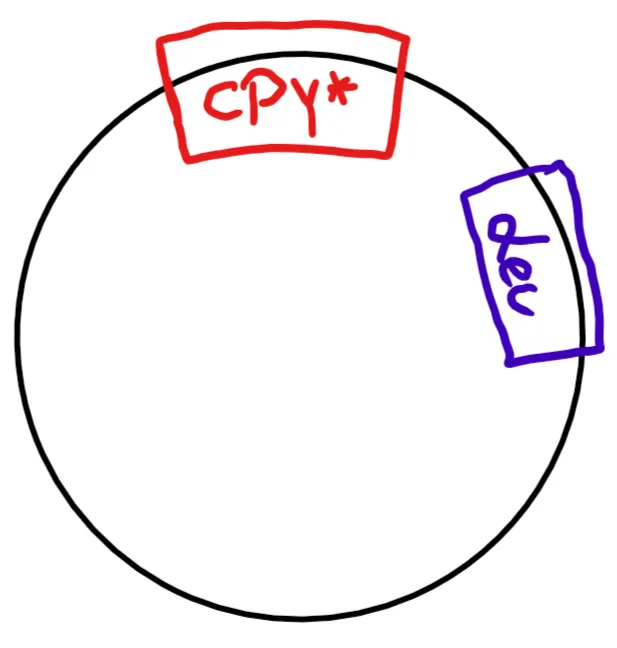
The real problem was CPY*. CPY* is the mutated version of a gene that usually codes for a protein destined for the secretory pathway. As such, it's perfect to monitor how a cell handles misfolded proteins.
In theory, you create a plasmid (round piece of DNA), which includes the gene you want (CPY*) and a so-called marker gene. A marker gene is supposed to tell you if the organism you're transforming (=inserting DNA into) absorbed the DNA or not. It can be almost anything, bacteria are usually transformed with a marker gene for antibiotic resistance.
For the yeast, we used a gene that allowed the organism to produce Leucine (an amino acid) by itself, thus enabling it to grow in a petri dish with nutritional agar missing Leucine (= Leucine dropout medium).
My first step was to take some E. coli, into which the plasmid had been transformed. E. coli is great for multiplying Plasmids, so you don't have to build a thousand of them by hand.
The E. coli grow overnight in a liquid medium, then the plasmids are isolated by destroying the cells (the process took about 45 minutes after I got used to doing it). Then, the purified plasmids are transformed into the yeast (1.5 to 2 hours, depending on how good my day was going) and the yeast can grow for 2-3 days on the dropout medium.
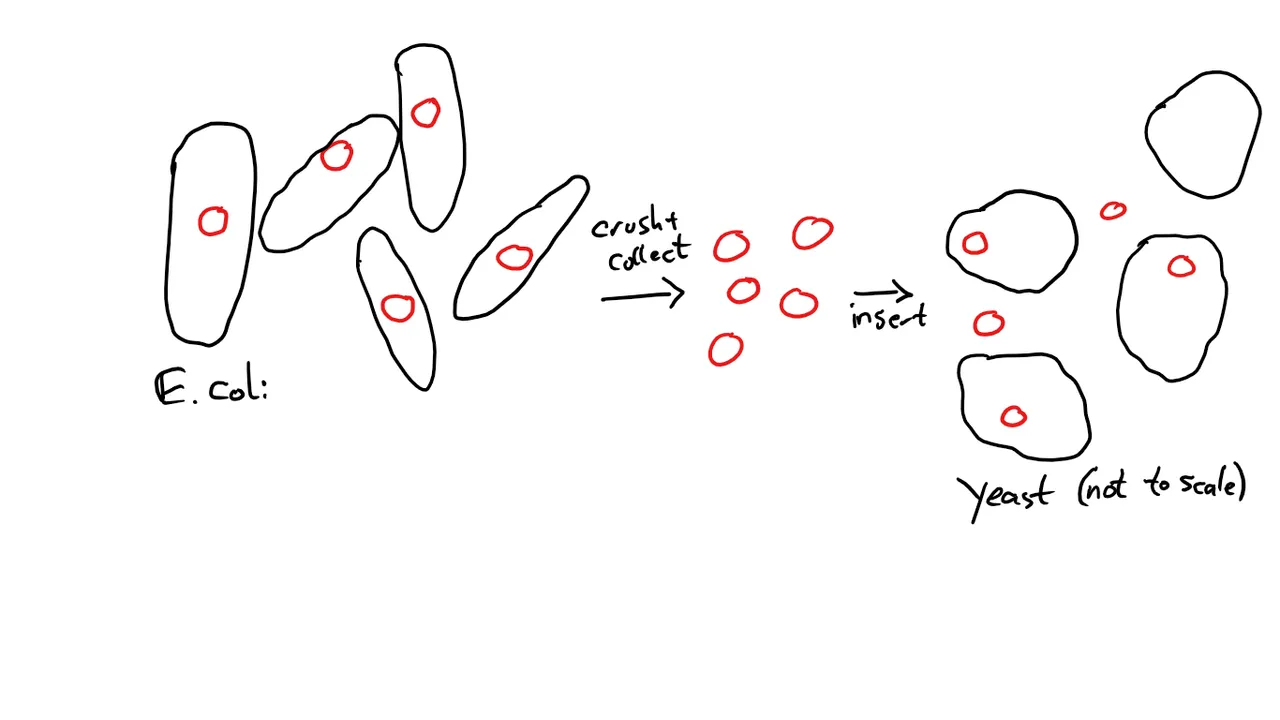
As you can see, not every yeast cell absorbed a plasmid during this process. That's why we need the marker gene and the dropout medium: We only want yeast cells that have been successfully transformed.
In theory, my plate with the yeast was supposed to look like this after 3 days:

The sad truth was ... nothing happened.
But I was a 4th-semester student with not a lot of lab experience, I had probably just done something wrong! Maybe the yeast hadn't properly absorbed the plasmid, after all, it was a foreign gene and yeast isn't so super happy about introducing foreign DNA into its cell.
I repeated the process.
And then I repeated it again.
And again.
At this point, my supervisor did the extraction of the plasmid and yeast transformation himself, to give me the chance to continue with my research project.
He failed too.
That was the point when we considered something might be off. 4 weeks had already passed of the 7 I had been given in that lab and we weren't even close to getting results!
To find out what might be wrong, we did a PCR to check if the gene had been successfully inserted.
This means we took some of the yeast cells and duplicated their DNA by heating it so it separates into two strands, which then were completed to new DNA strands, effectively doubling the amount of available Gene DNA.
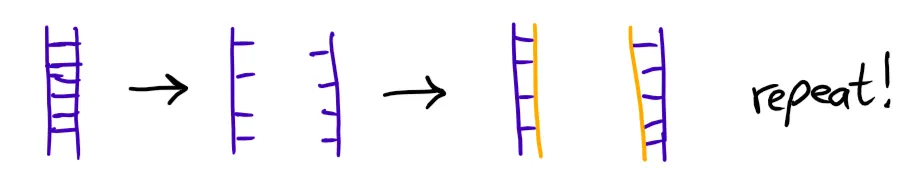
That's done several times (the whole thing took about 2 hours), then the DNA is put on a gel for a gel electrophoresis (please just read the Wikipedia entry).
The CPY* gene was there.
So we tried again.
And failed. Both.
So we repeated the PCR and sent some samples to a company, to have them sequence the plasmid and tell us if something is off.
The CPY* gene was there.
Our transformation didn't work.
I wonder if one of you guys already has an idea what might have gone wrong? We only noticed our mistake in week 6.
All the time, we had been checking if the CPY* gene was there. We never checked for the marker gene, because why should we? That's supposed to be pretty stable.
We did a PCR to check for the marker gene. It was not there. The person who had built this construct (another student, apparently) had managed to build a plasmid without marker gene and thus ruined 7 weeks of work for me.
That day, I sat at the computer in the lab, looking at the results and just thinking ...

I learned two things during these 7 weeks:
- When you're troubleshooting, check all possible ways something might have gone wrong, not just the most likely
- Do everything yourself. Trust no one. I could have built that plasmid myself, transformed the yeast AND done my testing in those 7 weeks. Instead, I was left with no results and slight disappointment.
Conclusion
As I said at the beginning, it's not really my shittiest "method", but most of my time in the lab has been spent with experiments tailored to teach students something, not to annoy them. I am sure there are a lot more shitty methods to come
As the next person, I am nominating @ertwro !
Pictures by me, except for the first one, that's from pixabay.com
