Hi friends... To be honest I am overwhelmed with many things recently. And few days back my tinkering became official work(Well... Kinda). Few of you might have seen my previous post. So I will elaborate a little bit more about how I started these projects, what am I intending to post, etc.
How I started this work?
I was applying for postdoctoral positions when I was about to wrap up my Ph.D. I was always fascinated by lipid membrane simulations. And, then I got selected as a postdoctoral researcher to a group that does coarse-grained lipid simulations, etc. So I was supposed to learn a new tool called LAMMPS software. Out of pure curiosity and free time I got during these COVID19 days I started working on replicating my postdoc supervisor's model in LAMMPS[1]. I extended it to vesicles too. You can see the details in the paper. Also, I am planning to write about these adventures in an upcoming series.
My upcoming plan for articles
- Description of the lipid model
- How to set up a coarse-grained vesicle using moltemplate/LAMMPS/topotools/VMD
- How to generate customized forcefield files for the simulations
- How to set up the simulations
- More details like analysis/interpretation of results and things according to how project evolves.
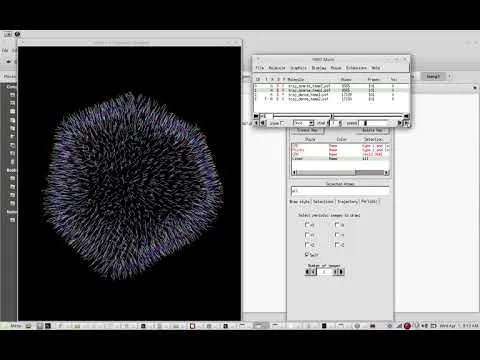
Screen Capture of simulations
Here is a screen capture video from my work. The coarse-grained lipid vesicle was simulated in LAMMPS under NVE ensemble and visualized in VMD software. I will explain the technical details in the upcoming articles.
Can you see the spherical vesicle getting a lot of sharp edges in the video? Physicists call them DEFECTS. These defects are assumed to form due to the rigidity in the lipid trimer. I will talk about them in the coming articles. @scienceangel : I don't know if you are seeing this. Do you remember our discussion about lipid flipflops? These simulations capture that phenomenon. I will show those too. Wait for it. :)
Till then bye bye!