Hello, my dear friends! Welcome to yet another science article from @dexterdev. Today I am going to write about something which I was thinking to do for the last few months. This is about my first research paper which got published in Nature Scientific Reports journal on August 28th this year. Due to my Ph.D. works, I got very busy in between and had to shift the plan of writing it up. I will be presenting this work and its extension as a poster for the Indo-US conference at ACTREC, Mumbai which will be happening from 7th to 11th December. So I thought this is the best time to write about our work. I will explain our work in the following sections:
- Background
- The Problem statement
- Simulation Technique used
- Results
- Summary
Background
So our main character in this work is a small GTPase protein called Rap protein. The main functions of Rap proteins are[1]:
- Cell proliferation
- Cell adhesion
- and other processes like cell junction formation
Rap proteins which belong to Ras-like superfamily are oncoproteins. Which means that certain mutations in these proteins can lead to cancer! The Ras/Ras-like proteins are players in the famous MAPK/ERK pathway.
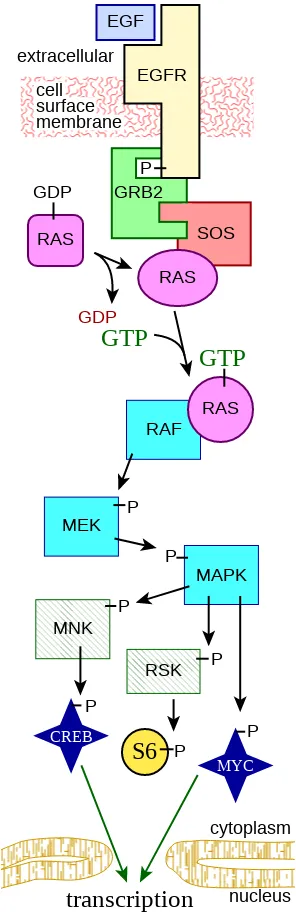
MAPK/ERK pathway. (Author: Fred the Oyster, Source: Wikimedia Commons, License: CC by SA ver 4.0)
MAPK/ERK signaling pathway takes signals from extracellular regions to the nucleus of the cell via different proteins. And the information maybe likes to execute cell division etc. But when mutations in the proteins involved in this particular pathway can cause cancer because sometimes the mutations can switch particular proteins in the cascade as permanently ON which implies the cell gets a signal to divide itself for indefinitely. These uncontrolled cell divisions can initiate cancer. Ras-like protein associated mutations are associated with 30% of all human cancers[2].
The image below shows Rap protein in the complexed state with another protein called Raf. The functional units in Rap is highlighted: P-loop, SWITCH-I, and SWITCH-II.
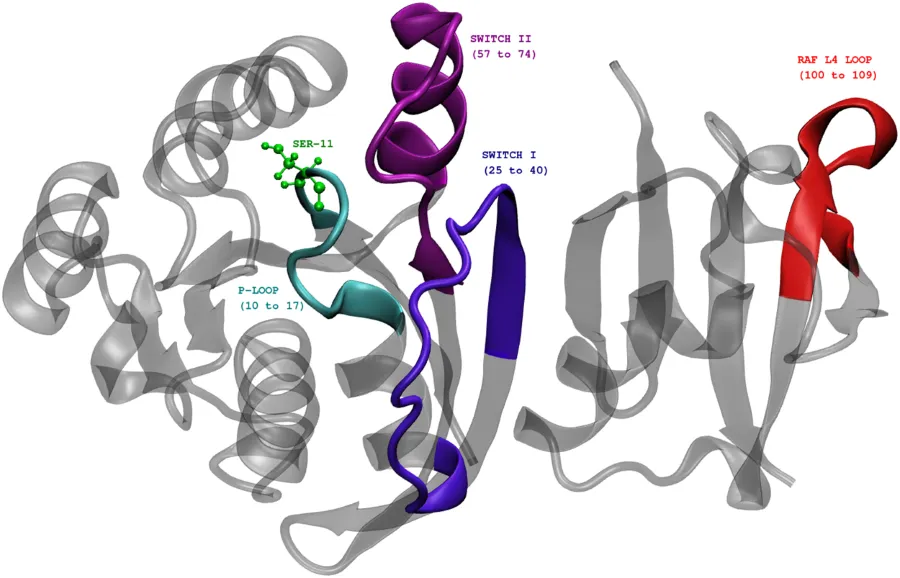
Rap protein complexed with Raf RBD(Ras binding domain). Ref:Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics. License:CC Ver 4. Author's own work.
Ras-like proteins act like a molecular switch. GTP bound states of these proteins are active and GDP bound states are inactive. Proteins called GEFs exchange GDP for GTP and GAPs accelerate(catalyse) the hydrolysis of GTPconverting it back to inactive GDP-bound protein.
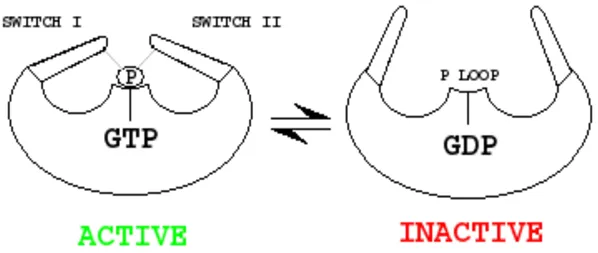
A cartoon representation of Ras-like protein states (My creation)
This video gives a very nice introduction to Ras proteins and their significance.
Youtube Video by Dustyn Levenson from the channel "University of Rochester Introductory Biochemistry (Bio250H)"
The Problem Statement
So it is widely known that Ras-like proteins are very hard to target using drug moelcules. (Mutations of Ras proteins at residues 12,13 and 61 can cause cancer) It is considered as one of the most undruggable molecules![3] People are currently looking into alternative methods to tame this molecule. One of the aspects people trying to see is that how do Post-translational modifications of Ras-like proteins affecting its behavior. Post-translational modifications a.k.a PTMs are reversible modifications caused to proteins(by other proteins) like phosphorylation or methylation etc. Phosphorylation is the addition of phosphate groups to residues in proteins which enables them to do certain tasks or changes the proteins' state. This is a nice regulation mechanism in proteins. People have found that single residue phosphorylation can really change the binding affinity between Ras-like proteins and its effector proteins through allosteric signal propagation in the protein complexes.[4,5,6,7] Allosteric signal propagation in short: It is a distal change which happens in proteins/protein-complexes as a result of minor changes like of mutation, PTM or upon binding with a moecule at a site. For example, a mutation at site one can bring a difference in the usual orientation of a loop very way. And this change is described using a analogy of domino-effect. A 26 second video about allostery here:
Reference: Youtube Channel Biotech Review
We were trying to see computationally the effects of phosphorylation of a single residue SER11(Serine 11). SER11 is a good candidate for experiments as it is in the functional P-loop region. SER11 phosphorylation was identified in Sharma, K. et al study[8].
Simulation Technique
We used All-atom Classical Molecular Dynamics(see my previous posts for articles regarding MD technique) as the technique to simulate the system. The Rap1A and its effector(cRaf1) RBD(Ras binding domain) structure was from protein data bank id 1C1Y[9]. The primary MD trajectories which we analysed were:
- GTP liganded Rap-Raf complex in a water box (400ns) [CODE in the paper: GTP-SER11 or simply GTP]
- GTP liganded Rap-Raf complex with Rap phosphorylated at SER11 in a water box (400ns) [CODE in the paper: GTP-PSER11]
The force field used was CHARMM36. We ran 400ns using NPT ensemble(Constant particles, constant pressure@1atm, constant temperature@298K).
Results
Here I will highlight some important results:
Allosteric signal transmission in Rap-RBD complex
To look into allosteric transmission we did community network analysis via NetworkView plugin in VMD. It uses Girvan-Newman algorithm on Rap-RBD complex. We found that GTP-PSER11 case exhibits connectedness from SER11 to loop L4 in Raf side. Yellow community is the L4 loop in the figure below. Also we see more communities in common to both protein chains in GTP-PSER11(phosphorylated) case.
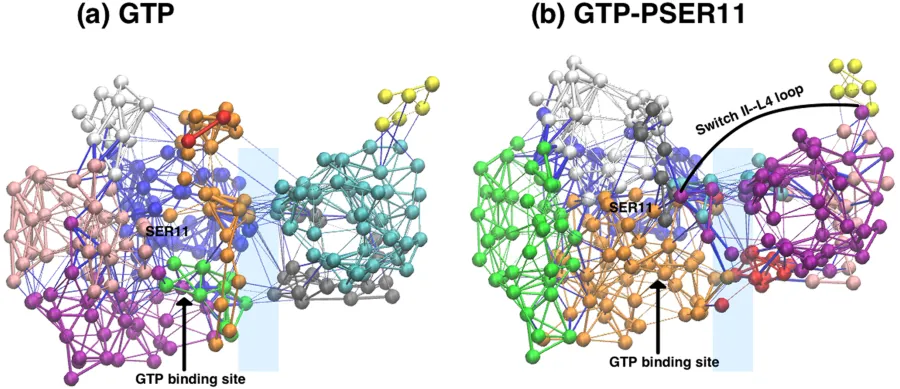
GTP-SER11 and GTP-PSER11(phosphorylated cases) Residue interaction networks. Ref:Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics. License:CC Ver 4. Author's own work.
Sampled Conformational space
The sampled conformational space is lesser in the case of Rap(Phosphorylated case). We did PCA(Principal Component Analysis) on the last 375-400ns MD data for this purpose. See the figure below:
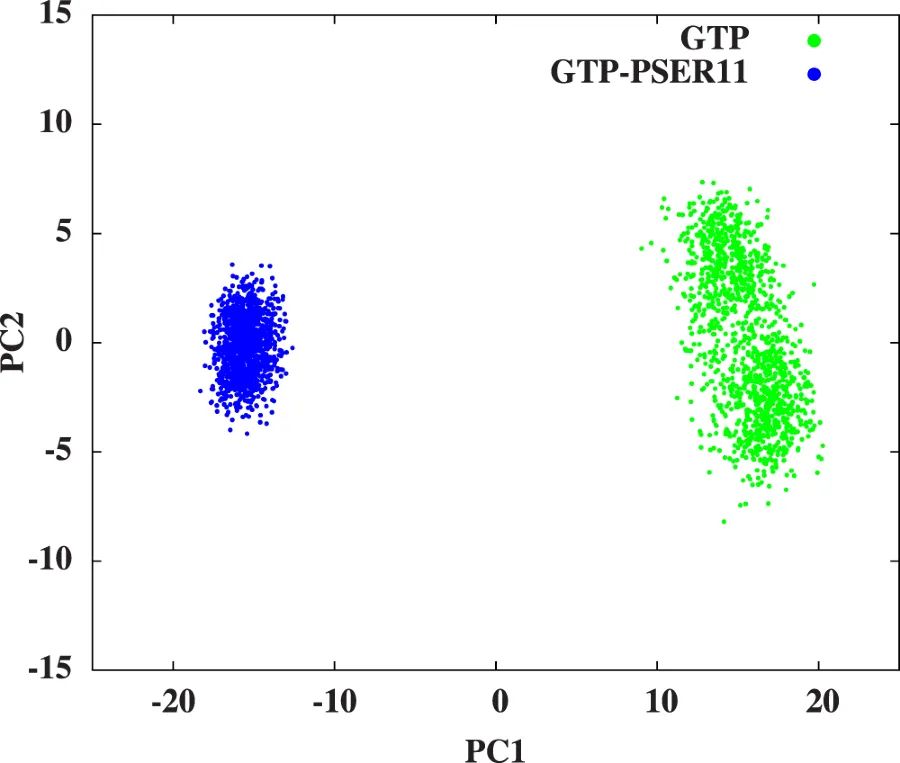
PCA done on both trajectories. Notice the smaller area for GTP-PSER11 case which indicates that Rap under SER11 phosphorylated case samples a smaller configurational space. It is more tighter! Ref:Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics. License:CC Ver 4. Author's own work.
(You may want to see how PCA is done: Link to article on how to do PCA.)
New pockets
We performed pocket analysis using CASTp online software[10]. Firstly the averaged Rap trajectories from 395 to 400ns data was considered and average structures were calculated. These averaged structures were supplied to CASp servers to find the biggest pockets/cavities in the proteins. We saw that GTP-PSER11 case had a differently located cavity. Also it was a smaller in volume. See the figure:
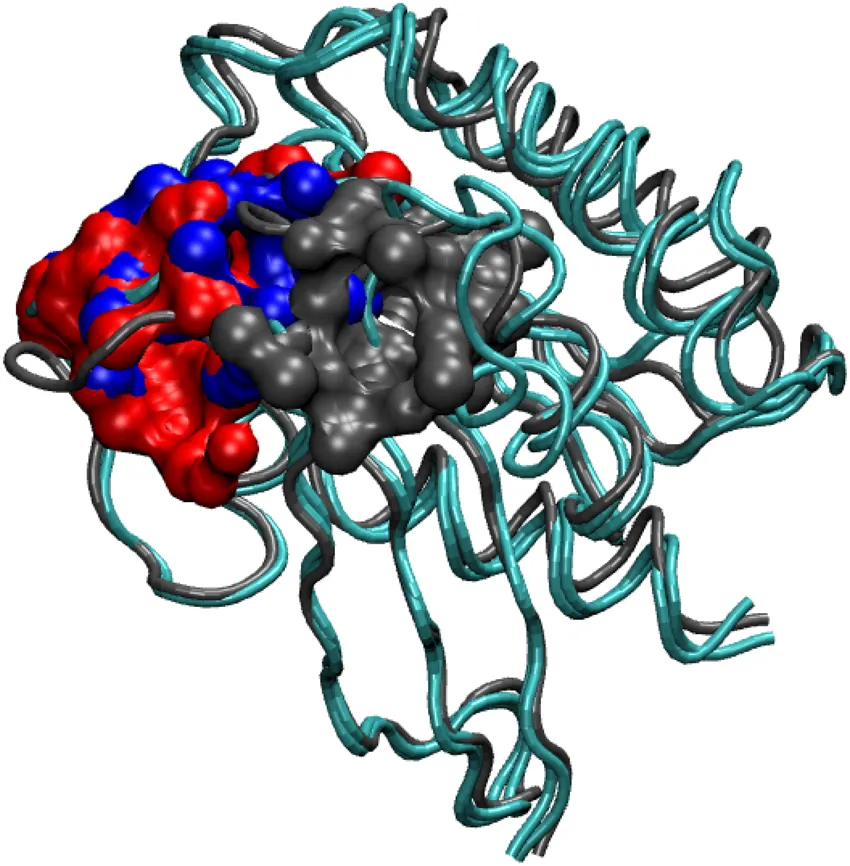
Black cavity: GTP-PSER11 case(smallest and different location), Blue cavity: Rap from 1C1Y crystal structure, Red cavity: GTP-SER11 case Ref:Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics. License:CC Ver 4. Author's own work.
Binding affinity
We also did the binding affinity free energy calculations using MM-GBSA method. (More on MM-GBSA techniques in a different article). The calculations suggest that SER11 phosphorylation improves binding between Rap and Raf-RBD.
Summary
The higher fluctuation dynamics nature of Rap structure(in nonphosphorylated case) around SER11 from simulations(via RMSF results) is indicative of the fact that SER11 can get phosphorylated possibly upon exposure from the buried situation. This is also confirmed from the fact that the SASA(solvent accessible surface area) values of SER11 in GTP-SER11 case is quite higher than in the crystal strucutre case. The main take away from this research work is that the SER11 phosphorylation stabilizes the Rap structure and is likely to stop GTP-hydrolysis which can ultimately lead the signal to reach L4 loop in RBD allosterically. The increased binding affinity between Rap and Raf upon SER11 phosphorylation maybe a result of improved electrostatic interactions. Also, PCA results show that the Rap chain in GTP-PSER11 case samples a lesser conformational space, which is indicative of the fact Rap is in a "tight" state. One of the functions of Rap protein is to competitively bind to Raf(compared to Ras) and keep it inactive and silence the Ras-Raf binding. Ras has a crucial GLN61 which plays a great role in GTP hydrolysis, whereas in Rap this is replaced by THR61 which doesn't take part in hydrolysis. Upon SER11 phosphorylation it is seen that the Rap binding with Raf-RBD becomes more strong which makes the interaction of any Ras with Raf molecules rare. But Whether this improved binding of phosphorylated Rap wth Raf-RBD can initiate changes in Raf-CRD(Cysteine rich domain), which inturn is necessary to signal transmission in the MAPK/ERK pathway is still under speculation.
We think that this study is a great example of utilizing computational tools to probe a plethora of hidden information in "static" crystal/NMR structures. We also believe that one may target a kinase for phosphorylation of notoriously druggable proteins such as Ras-family proteins.
References
- [0] Our work: Devanand, T., Prasanna Venkatraman, and Satyavani Vemparala. "Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics." Scientific reports 8.1 (2018): 12976.
- [1] https://www.ncbi.nlm.nih.gov/gene/5906
- [2] Lu, S., Jang, H., Gu, S., Zhang, J. & Nussinov, R. Drugging Ras GTPase: A comprehensive mechanistic and signaling structural view. Chem Soc Rev 45, 4929–4952 (2016).
- [3] Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J. & Der, C. J. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13, 828 (2014).
- [4] Ting, P. Y. et al. Tyrosine phosphorylation of RAS by ABL allosterically enhances effector binding. FASEB J 29, 3750–3761 (2015).
- [5] Fremin, C. et al. ERK1/2-induced phosphorylation of R-Ras GTPases stimulates their oncogenic potential. Oncogene 35, 5692–5698 (2016).
- [6] Sahyoun, N., McDonald, O. B., Farrell, F. & Lapetina, E. G. Phosphorylation of a Ras-related GTP-binding protein, Rap-1b, by a neuronal Ca2+/calmodulin-dependent protein kinase, CaM kinase Gr. Proc Natl Acad Sci USA 88, 2643–2647 (1991).
- [7] Bunda, S. et al. Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation. Proc Natl Acad Sci USA 111, E3785–E3794 (2014).
- [8] Sharma, K. et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep 8, 1583–1594 (2014).
- [9] Nassar, N., Horn, G., Herrmann, C., Scherer, A. et al. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with RaplA and a GTP analogue. Nature 375, 554 (1995).
- [10] Dundas, J. et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34, W116–W118 (2006).
#steemSTEM
#steemSTEM is a very vibrant community on top of STEEM blockchain for Science, Technology, Engineering and Mathematics (STEM). If you wish to support steemstem visit the links below:

Quick link for voting for the SteemSTEM Witness(@stem.witness)
Delegation links for @steemstem gives ROI of 65% of curation rewards
(quick delegation links: 50SP | 100SP | 500SP | 1000SP | 5000SP | 10000SP).
Also visit the steemstem app here: https://www.steemstem.io
Follow me @dexterdev
____ _______ ______ _________ ____ ______
/ _ / __\ \//__ __/ __/ __/ _ / __/ \ |\
| | \| \ \ / / \ | \ | \/| | \| \ | | //
| |_/| /_ / \ | | | /_| | |_/| /_| \//
\____\____/__/\\ \_/ \____\_/\_\____\____\__/

credit: @mathowl
So bye bye friends. See you people with another interesting article. Till then STEEM ON!