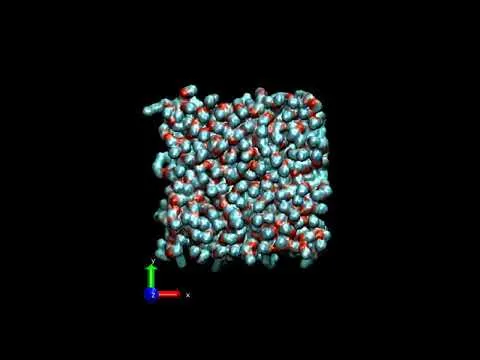
Recently we did some all-atom classical molecular dynamics simulations involving DMPC model membrane, Nogo66 membrane active protein, TIP3P water model and NaCl ions. We used CHARMM36 forcefield and ran the simulations in NAMD software. VMD was the software used to visualize system. (If you are new to these, I have written many previous articles relating to all things which I have just mentioned. Please feel free to dig my previous articles. I will give some links as references.)
Lipid defects
Lipid molecules which form cellular membranes and membranes of other oragnelles are amphipathic (a.k.a amphiphilic) molecules which means they have both hydrophobic(water hating) and hydrophilic(water liking) regions.

The fatty acid legs are hydrophobic,and the head region(other than tail) is hydrophilic. From one of my old articles. Link here
Lipid defects are those regions in bilayers where the hydrophobic tails get exposed to water, which is energetically very unfavourable. Defects can get get induced by many factors. One being interaction of membrane with particular molecules(I will write more about lipid defects in a future article.)
Defects can lead to lipid interdigitation. Means the lipid tails in both leaflets can overlap as shown below:
The state with lipid defects is energetically unfavourable. So lipids reorganize themselves to a favourable interdigitated state.[1]
So below is a youtube video which I created. This shows defects:
This is a 300ns all-atom classical molecular dynamics simulation of Nogo66 protein (from pdb structure 2KO2) on a model DMPC bilayer patch (144 lipids in each leaflet). This setup is solvated in TIP3P water model + 0.15 M NaCl ion concentration. Only the top monolayer in the bilayer is shown for clearly visualizing the lipid defects. The black voids are defects. Look for it. If you feel the video is very fast, please reduce the speed to 0.5 or so in the youtube settings.
References
Research paper:
My previous posts:
To learn about VMD and PDB file format, see here:
- https://www.steemstem.io/#!/@dexterdev/visualizing-bio-molecules-in-computer-part-1-let-us-inspect-a-pdb-file-and-see-it-using-vmd
- https://www.steemstem.io/#!/@dexterdev/visualizing-bio-molecules-in-computer-part-2-introduction-to-tcl-scripting-environment-in-vmd-1-sbd-prize-task-inside
To learn about the concepts in All-atom molecular dynamics see articles below:
- https://www.steemstem.io/#!/@dexterdev/classical-molecular-dynamics-series-part-1-the-fundamentals
- https://www.steemstem.io/#!/@dexterdev/classical-molecular-dynamics-series-part-2-the-force-field
- https://www.steemstem.io/#!/@dexterdev/classical-molecular-dynamics-series-part-3-solving-the-molecular-dynamics-equation
To setup and run simulations in NAMD software, see below:
- https://www.steemstem.io/#!/@dexterdev/classical-molecular-dynamics-series-part-4a-let-us-setup-a-simulation-and-run-it
- https://www.steemstem.io/#!/@dexterdev/classical-molecular-dynamics-series-part-4b-running-small-systems-on-your-computer
- https://www.steemstem.io/#!/@dexterdev/let-us-cool-dmpc-bilayer-lipids-an-18-day-long-molecular-dynamics-experiment-on-hpc-facility
Textbook references for learning theory of Molecular Dynamics:
- "Statistical Mechanics: Theory and Molecular Simulations" by Mark E. Tuckerman
- "Molecular Modelling: Principles and Applications" by Andrew R. Leach
- "Computer Simulation of Liquids" by D. J. Tildesley and M.P. Allen
References specific to NAMD and VMD:
#steemSTEM
#steemSTEM is a very vibrant community on top of STEEM blockchain for Science, Technology, Engineering and Mathematics (STEM). If you wish to support steemstem visit the links below:

Quick link for voting for the SteemSTEM Witness(@stem.witness)
Delegation links for @steemstem give ROI of 65% of curation rewards
(quick delegation links: 50SP | 100SP | 500SP | 1000SP | 5000SP | 10000SP).
Also visit and create your STEM posts using steemstem app here: https://www.steemstem.io
You can also set @steemstem as beneficiary.
Follow me @dexterdev
____ _______ ______ _________ ____ ______
/ _ / __\ \//__ __/ __/ __/ _ / __/ \ |\
| | \| \ \ / / \ | \ | \/| | \| \ | | //
| |_/| /_ / \ | | | /_| | |_/| /_| \//
\____\____/__/\\ \_/ \____\_/\_\____\____\__/

credit: @mathowl
Signing off now! I will be back with some interesting topic from my area of research soon.