Hi friends...
Recently I had written about our research work which got published in this August. Those who have not read that article can find it below:
Phosphorylation promotes binding affinity of Rap-Raf complex [Original Research Work Summary]
Those who were following me here on steem blockchain might be knowing that I was presenting this work in form of a poster presentation at ACTREC, Mumbai. One of the frequent questions which we have encountered is why Serine11(SER11) residue was chosen as the site for phosphorylation study using Molecular dynamics. So in this small article, I will address this question.
SER11 lies in P-loop
P-loop (phosphate-binding loop, 10-17 residue positions) is one of the most functionally important loops in Ras-like GTPase proteins. It is one of the conserved motifs in ATP and GTP-binding proteins. The P-loop is a glycine-rich loop followed by a lysine and Serine/Threonine residues[1]. In the case of GTPases, the beta and gamma phosphates of GTP interacts strongly with P-loop[2]. Certain mutations in P-loop like at cites Glycine-12 and Glycine-13 can be oncogenic[1].
Since Serine11 is in P-loop which is a functionally important loop, it would be nice to examine the perturbations we can induce on this particular loop. That is one of the reasons why we chose this specific site in this Rap1A system.
The image below shows Rap protein in the complexed state with another protein called Raf. The functional units in Rap are highlighted: P-loop, SWITCH-I, and SWITCH-II. Also highlights SER11.
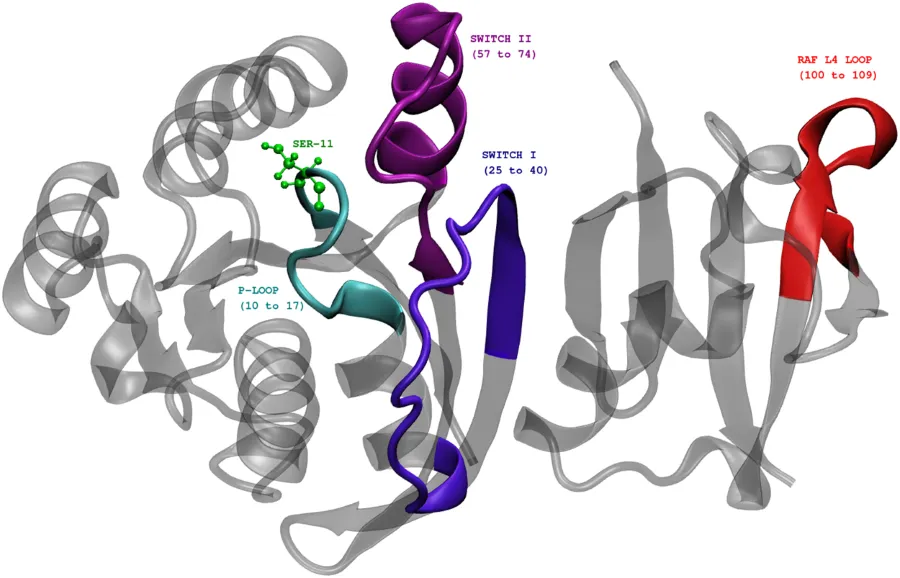
Rap protein complexed with Raf RBD(Ras binding domain). Ref:Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics. License:CC Ver 4. Author's own work.
SER11 phosphorylations have been reported in past
SER11 phosphorylation events have been reported in previous experiments[3]. Sharma et al[3] have reported SER11, SER39 phosphorylations in Rap1A protein.
Also, this paper reports that phosphorylation events in proteins follows a 80/20 rule also popularly known as pareto distribution(Power law). This implies that most of the proteins undergo very less number of phosphorylations. But this is an entire topic altogether.
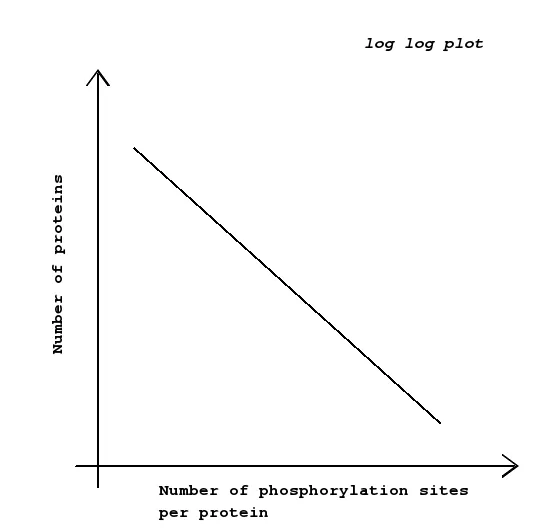
The 80/20 rule in protein phosphorylations(Post translational modifications in general). This plot implies that most of the proteins will experience very few phosphorylations(1 or 2). (Power law has other implications too. A subject for maybe another article.) :)
Phosphorylation requires exposure!
This section requires the understanding of solvent accessible surface area a.k.a SASA. SASA of a molecule is its surface area accessible to solvent. Say a protein is in water. You can then try measuring the SASA of that protein. You can also measure SASA of a particular region in a protein like a single residue etc.
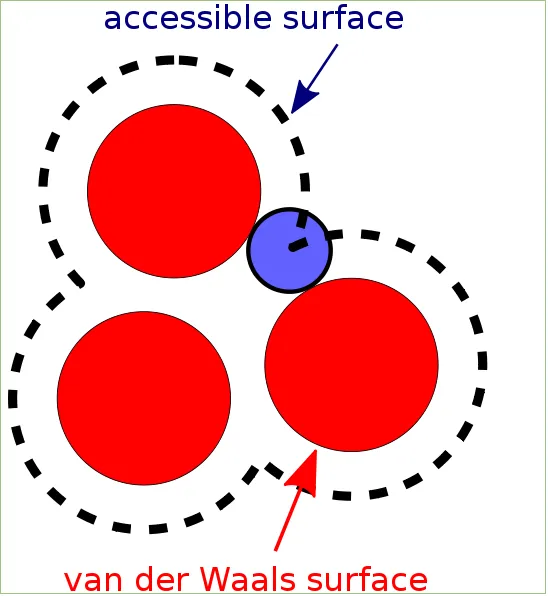
SASA Illustration
Author: Keith Callenberg, Source: Wikimedia Commons, License: Creative Commons Attribution-Share Alike 3.0 Unported
We know at least in many cases crystal structure shows residues like SER11 in P-loop with lesser solvent accessiblity. So we were curious to look at its dynamics using Molecular Dynamics simulations. The figure below make things clear. This is from our Supporting documents to the paper.
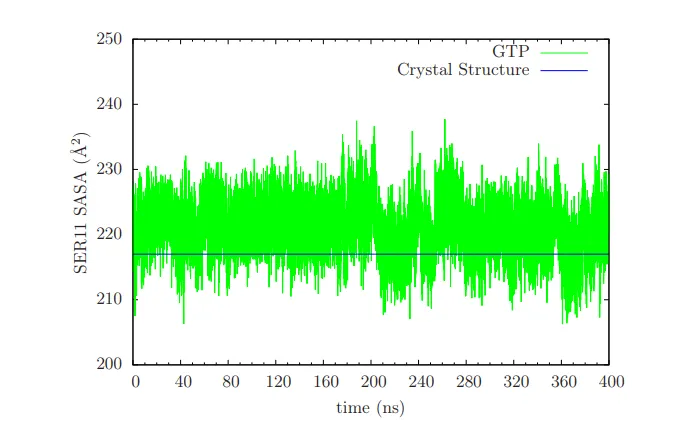
The solvent accessible surface area(SASA) for the crystal structure case is like 218 Angstrom^2. You can see that in the time course of the simulation, the SER11 residue gets exposed more. Which makes it more probable to get phosphorylated by the related kinase protein.
Conclusion
As mentioned in the previous article, we found that the binding affinity of Rap-Raf complex improves as a consequence of phosphorylation of SER11 residue(as suggested by the MD simulations). We saw that why SER11 residue is significant. We hope that this MD study will initiate experiments to poke into the hypothesis of phosphorylation can accelerate the Rap1A competitveness in binding with Raf comparing to Ras proteins.
References
- [0] Our work: Devanand, T., Prasanna Venkatraman, and Satyavani Vemparala. "Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics." Scientific reports 8.1 (2018): 12976.
- [1] Saraste, Matti, Peter R. Sibbald, and Alfred Wittinghofer. "The P-loop—a common motif in ATP-and GTP-binding proteins." Trends in biochemical sciences 15.11 (1990): 430-434.
- [2] Vetter, Ingrid R., and Alfred Wittinghofer. "The guanine nucleotide-binding switch in three dimensions." Science 294.5545 (2001): 1299-1304.
- [3] Sharma, Kirti, et al. "Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling." Cell reports 8.5 (2014): 1583-1594.
#steemSTEM
#steemSTEM is a very vibrant community on top of STEEM blockchain for Science, Technology, Engineering and Mathematics (STEM). If you wish to support steemstem visit the links below:

Quick link for voting for the SteemSTEM Witness(@stem.witness)
Delegation links for @steemstem gives ROI of 65% of curation rewards
(quick delegation links: 50SP | 100SP | 500SP | 1000SP | 5000SP | 10000SP).
Also visit the steemstem app here: https://www.steemstem.io
Follow me @dexterdev
____ _______ ______ _________ ____ ______
/ _ / __\ \//__ __/ __/ __/ _ / __/ \ |\
| | \| \ \ / / \ | \ | \/| | \| \ | | //
| |_/| /_ / \ | | | /_| | |_/| /_| \//
\____\____/__/\\ \_/ \____\_/\_\____\____\__/

credit: @mathowl